PubMed API in Python#
by Avery Fernandez
The PubMed API, part of NCBI’s Entrez Programming Utilities (E-utilities), provides programmatic access to biomedical literature from the PubMed database, enabling retrieval of bibliographic data.
This tutorial content is intended to help facilitate academic research.
Please see the following resources for more information on API usage:
Documentation
Terms
Data Reuse
NOTE: The PubMed API (Entrez E-utilities) limits requests to a maximum of 3 requests per second without an API key, and up to 10 requests per second with an API key.
These recipe examples were tested on April 9, 2025.
Setup#
The following external libraries need to be installed into your environment to run the code examples in this tutorial:
We import the libraries used in this tutorial below:
from time import sleep
import requests
from pprint import pprint
import matplotlib.pyplot as plt
from datetime import datetime
1. Retieve the Metadata of an Article#
The article we are requesting has PubMed ID: 27933103
retmode in the web API URL specifies the file format. In this example, we use JSON.
ESUMMARY_URL = "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esummary.fcgi"
params = {
'db': 'pubmed',
'id': '27933103',
'retmode': 'json'
}
try:
response = requests.get(ESUMMARY_URL, params=params)
# Raise an error for bad responses
response.raise_for_status()
data = response.json()
pprint(data, depth=3)
except requests.exceptions.RequestException as e:
print(f"An error occurred: {e}")
data = None
{'header': {'type': 'esummary', 'version': '0.3'},
'result': {'27933103': {'articleids': [...],
'attributes': [...],
'authors': [...],
'availablefromurl': '',
'bookname': '',
'booktitle': '',
'chapter': '',
'doccontriblist': [],
'docdate': '',
'doctype': 'citation',
'edition': '',
'elocationid': '',
'epubdate': '2016 Nov 23',
'essn': '1758-2946',
'fulljournalname': 'Journal of cheminformatics',
'history': [...],
'issn': '1758-2946',
'issue': '',
'lang': [...],
'lastauthor': 'Bara JE',
'locationlabel': '',
'medium': '',
'nlmuniqueid': '101516718',
'pages': '66',
'pmcrefcount': 33,
'pubdate': '2016',
'publisherlocation': '',
'publishername': '',
'pubstatus': '258',
'pubtype': [...],
'recordstatus': 'PubMed',
'references': [],
'reportnumber': '',
'sortfirstauthor': 'Scalfani VF',
'sortpubdate': '2016/11/23 00:00',
'sorttitle': 'programmatic conversion of crystal '
'structures into 3d printable files '
'using jmol',
'source': 'J Cheminform',
'srccontriblist': [],
'srcdate': '',
'title': 'Programmatic conversion of crystal '
'structures into 3D printable files using '
'Jmol.',
'uid': '27933103',
'vernaculartitle': '',
'volume': '8'},
'uids': ['27933103']}}
Data extraction: Let us try to extract the authors of the paper
# Check if we got data back from the API
if data:
# Grab the list of authors from the response
authors = data["result"]["27933103"]["authors"]
# Loop through the authors and print their names
for author in authors:
print(f"{author['name']}")
Scalfani VF
Williams AJ
Tkachenko V
Karapetyan K
Pshenichnov A
Hanson RM
Liddie JM
Bara JE
2. Retrieve Metadata for a List of PubMed IDs#
First, create a list of PubMed IDs:
ids = [34813985, 34813932, 34813684, 34813661, 34813372, 34813140, 34813072]
Now we can go about acquiring the data from PubMed and saving the data in a dictionary, called multi_papers:
multi_papers = {}
for pubmed_id in ids:
params = {
'db': 'pubmed',
'id': pubmed_id,
'retmode': 'json'
}
try:
response = requests.get(ESUMMARY_URL, params=params)
# Add a delay between API calls
sleep(.5)
# Raise an error for bad responses
response.raise_for_status()
data = response.json()
multi_papers[pubmed_id] = data
except requests.exceptions.RequestException as e:
print(f"An error occurred for ID {pubmed_id}: {e}")
# View first result
pprint(multi_papers[ids[0]], depth=3)
{'header': {'type': 'esummary', 'version': '0.3'},
'result': {'34813985': {'articleids': [...],
'attributes': [...],
'authors': [...],
'availablefromurl': '',
'bookname': '',
'booktitle': '',
'chapter': '',
'doccontriblist': [],
'docdate': '',
'doctype': 'citation',
'edition': '',
'elocationid': 'doi: 10.1016/j.ceca.2021.102500',
'epubdate': '2021 Nov 8',
'essn': '1532-1991',
'fulljournalname': 'Cell calcium',
'history': [...],
'issn': '0143-4160',
'issue': '',
'lang': [...],
'lastauthor': 'Morad M',
'locationlabel': '',
'medium': '',
'nlmuniqueid': '8006226',
'pages': '102500',
'pmcrefcount': 69,
'pubdate': '2022 Jan',
'publisherlocation': '',
'publishername': '',
'pubstatus': '256',
'pubtype': [...],
'recordstatus': 'PubMed - indexed for MEDLINE',
'references': [],
'reportnumber': '',
'sortfirstauthor': 'Fernández-Morales JC',
'sortpubdate': '2022/01/01 00:00',
'sorttitle': 'mutation in ryr2 fkbp binding site '
'alters ca 2 signaling modestly but '
'increases arrhythmogenesis in human '
'stem cells derived cardiomyocytes',
'source': 'Cell Calcium',
'srccontriblist': [],
'srcdate': '',
'title': 'Mutation in RyR2-FKBP Binding site alters '
'Ca(2+) signaling modestly but increases '
'"arrhythmogenesis" in human stem cells '
'derived cardiomyocytes.',
'uid': '34813985',
'vernaculartitle': '',
'volume': '101'},
'uids': ['34813985']}}
# Get title for each journal
for pubmed_id in ids:
pprint(multi_papers[pubmed_id]["result"][str(pubmed_id)]["source"])
'Cell Calcium'
'Methods'
'FEBS J'
'Dev Growth Differ'
'CRISPR J'
'Chembiochem'
'Methods Mol Biol'
3. PubMed API Calls with Requests & Parameters#
When searching through the articles, we are given a few of ways of filtering the data.
A list of all the available parameters for these requests can be found in the NCBI documentation.
We use the PubMed database:
db=<database>
We can, for example, use a query to search PubMed, such as “neuroscience intervention learning”:
term=<searchQuery>
ESEARCH_URL = "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi"
params = {
'db': 'pubmed',
'term': 'neuroscience intervention learning',
'retmode': 'json'
}
try:
response = requests.get(ESEARCH_URL, params=params)
# Raise an error for bad responses
response.raise_for_status()
data = response.json()
pprint(data, depth=3)
except requests.exceptions.RequestException as e:
print(f"An error occurred: {e}")
data = None
{'esearchresult': {'count': '30736',
'idlist': ['40249488',
'40249484',
'40248861',
'40247778',
'40247367',
'40247305',
'40245320',
'40245081',
'40244858',
'40242924',
'40240815',
'40240239',
'40239200',
'40239043',
'40238728',
'40238239',
'40236290',
'40235960',
'40233091',
'40232751'],
'querytranslation': '("neuroscience s"[All Fields] OR '
'"neurosciences"[MeSH Terms] OR '
'"neurosciences"[All Fields] OR '
'"neuroscience"[All Fields]) AND '
'("intervention s"[All Fields] OR '
'"interventions"[All Fields] OR '
'"interventive"[All Fields] OR '
'"methods"[MeSH Terms] OR "methods"[All '
'Fields] OR "intervention"[All Fields] '
'OR "interventional"[All Fields]) AND '
'("learning"[MeSH Terms] OR '
'"learning"[All Fields] OR "learn"[All '
'Fields] OR "learned"[All Fields] OR '
'"learning s"[All Fields] OR '
'"learnings"[All Fields] OR '
'"learns"[All Fields])',
'retmax': '20',
'retstart': '0',
'translationset': [{...}, {...}, {...}]},
'header': {'type': 'esearch', 'version': '0.3'}}
The number of returned IDs can be adjusted with the retmax paramater:
params = {
'db': 'pubmed',
'term': 'neuroscience intervention learning',
'retmode': 'json',
'retmax': 25
}
try:
response = requests.get(ESEARCH_URL, params=params)
# Raise an error for bad responses
response.raise_for_status()
data = response.json()
pprint(data, depth=2)
except requests.exceptions.RequestException as e:
print(f"An error occurred: {e}")
data = None
{'esearchresult': {'count': '30736',
'idlist': [...],
'querytranslation': '("neuroscience s"[All Fields] OR '
'"neurosciences"[MeSH Terms] OR '
'"neurosciences"[All Fields] OR '
'"neuroscience"[All Fields]) AND '
'("intervention s"[All Fields] OR '
'"interventions"[All Fields] OR '
'"interventive"[All Fields] OR '
'"methods"[MeSH Terms] OR "methods"[All '
'Fields] OR "intervention"[All Fields] '
'OR "interventional"[All Fields]) AND '
'("learning"[MeSH Terms] OR '
'"learning"[All Fields] OR "learn"[All '
'Fields] OR "learned"[All Fields] OR '
'"learning s"[All Fields] OR '
'"learnings"[All Fields] OR '
'"learns"[All Fields])',
'retmax': '25',
'retstart': '0',
'translationset': [...]},
'header': {'type': 'esearch', 'version': '0.3'}}
if data:
pprint(data["esearchresult"]["idlist"])
['40249488',
'40249484',
'40248861',
'40247778',
'40247367',
'40247305',
'40245320',
'40245081',
'40244858',
'40242924',
'40240815',
'40240239',
'40239200',
'40239043',
'40238728',
'40238239',
'40236290',
'40235960',
'40233091',
'40232751',
'40232556',
'40231304',
'40230768',
'40230302',
'40229794']
if data:
print(len(data["esearchresult"]["idlist"]))
25
We can also use the query to search for an author.
We add [au] after the name to specify it is an author.
params = {
'db': 'pubmed',
'term': 'Darwin[au]',
'retmode': 'json',
}
try:
response = requests.get(ESEARCH_URL, params=params)
# Raise an error for bad responses
response.raise_for_status()
data = response.json()
print(data["esearchresult"]["count"])
except requests.exceptions.RequestException as e:
print(f"An error occurred: {e}")
data = None
685
Sorting Results#
We can use the following to store the data for it to be sorted in the same API call.
usehistory=y
This will sort the ids by the publishing date:
sort=pub+date
params = {
'db': 'pubmed',
'term': 'Coral Reefs',
'retmode': 'json',
'usehistory': 'y',
'sort': 'pub date'
}
try:
response = requests.get(ESEARCH_URL, params=params)
# Raise an error for bad responses
response.raise_for_status()
data = response.json()
pprint(data["esearchresult"]["idlist"])
except requests.exceptions.RequestException as e:
print(f"An error occurred: {e}")
data = None
An error occurred: 429 Client Error: Too Many Requests for url: https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=pubmed&term=Coral+Reefs&retmode=json&usehistory=y&sort=pub+date
# Compare to unsorted
params = {
'db': 'pubmed',
'term': 'Coral Reefs',
'retmode': 'json',
}
try:
response = requests.get(ESEARCH_URL, params=params)
# Raise an error for bad responses
response.raise_for_status()
data = response.json()
pprint(data["esearchresult"]["idlist"])
except requests.exceptions.RequestException as e:
print(f"An error occurred: {e}")
data = None
An error occurred: 429 Client Error: Too Many Requests for url: https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=pubmed&term=Coral+Reefs&retmode=json
Searching Based on Publication Types#
We can sort by publication type by adding AND into the search:
term=<searchQuery>+AND+filter[filterType]
[pt] specifies that the filter type is publication type. More filters can be found at PubMed Help.
params = {
'db': 'pubmed',
'term': 'stem cells AND clinical trial[pt]',
'retmode': 'json',
}
try:
response = requests.get(ESEARCH_URL, params=params)
# Raise an error for bad responses
response.raise_for_status()
data = response.json()
pprint(data, depth=3)
except requests.exceptions.RequestException as e:
print(f"An error occurred: {e}")
data = None
An error occurred: 429 Client Error: Too Many Requests for url: https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=pubmed&term=stem+cells+AND+clinical+trial%5Bpt%5D&retmode=json
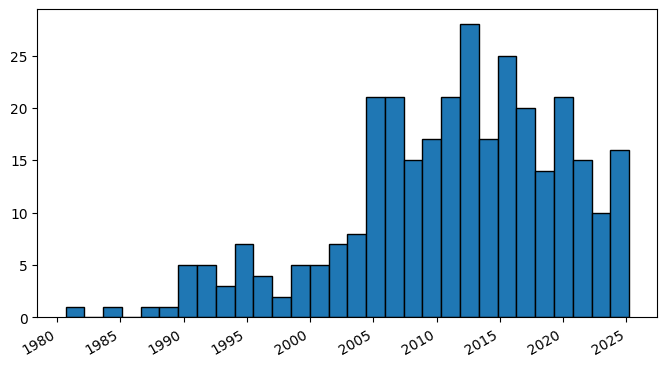
4. PubMed API metadata visualization#
Frequency of Topic sortpubdate Field#
Extracting the sortpubdate field for a “hydrogel drug” search results, limited to publication type clinical trials:
params = {
'db': 'pubmed',
'term': 'hydrogel drug AND clinical trial[pt]',
'retmode': 'json',
'usehistory': 'y',
'sort': 'pub date',
'retmax': 500
}
try:
response = requests.get(ESEARCH_URL, params=params)
# Raise an error for bad responses
response.raise_for_status()
data = response.json()
ids = data["esearchresult"]["idlist"]
pprint(ids[:10])
except requests.exceptions.RequestException as e:
print(f"An error occurred: {e}")
data = None
ids = []
['40119971',
'39937340',
'39904726',
'40000087',
'39825943',
'39384437',
'39147219',
'38971179',
'38932659',
'38875184']
len(ids)
316
# Loop through each IDs and get the sortpubdate field.
# Note that this sortpubdate field may not necassarily be equivalent to a publication date
pub_dates = []
for id in ids:
params = {
'db': 'pubmed',
'id': id,
'retmode': 'json'
}
try:
response = requests.get(ESUMMARY_URL, params=params)
sleep(.5)
# Raise an error for bad responses
response.raise_for_status()
data = response.json()
pub_dates.append(data["result"][str(id)]["sortpubdate"][0:10])
except requests.exceptions.RequestException as e:
print(f"An error occurred: {e}")
data = None
pprint(pub_dates[:10])
['2025/03/22',
'2025/03/01',
'2025/03/01',
'2025/02/25',
'2025/01/18',
'2024/11/01',
'2024/11/01',
'2024/10/01',
'2024/10/01',
'2024/10/01']
len(pub_dates)
316
Now that we have sortpubdates, if we want to visualize them in matplotlib, we have to convert it over to something it understands
matplotlib_dates = []
for date in pub_dates:
# Convert to datetime object
date = datetime.strptime(date, '%Y/%m/%d')
# Convert to matplotlib date format
matplotlib_dates.append(date)
pprint(matplotlib_dates[0:10])
[datetime.datetime(2025, 3, 22, 0, 0),
datetime.datetime(2025, 3, 1, 0, 0),
datetime.datetime(2025, 3, 1, 0, 0),
datetime.datetime(2025, 2, 25, 0, 0),
datetime.datetime(2025, 1, 18, 0, 0),
datetime.datetime(2024, 11, 1, 0, 0),
datetime.datetime(2024, 11, 1, 0, 0),
datetime.datetime(2024, 10, 1, 0, 0),
datetime.datetime(2024, 10, 1, 0, 0),
datetime.datetime(2024, 10, 1, 0, 0)]
fig, ax = plt.subplots()
plt.hist(matplotlib_dates, bins=30,edgecolor='black')
# set_size specifies the size of the graph
fig.set_size_inches(8, 4)
# Rotate and right-align the x labels so they don't crowd each other
for label in ax.get_xticklabels(which='major'):
label.set(rotation=30, horizontalalignment='right')
plt.show()