CAS Common Chemistry API in Python#
By Vincent F. Scalfani, Avery Fernandez, and Michael T. Moen
The CAS Common Chemistry API provides programmatic access to a curated subset, offering information on nearly 500,000 chemical substances.
Please see the following resources for more information on API usage:
Documentation
Terms
Data Reuse
CAS Common Chemistry is provided under the Creative Commons CC BY-NC 4.0 license
NOTE: The CAS Common Chemistry API limits requests to a maximum of 50 results per page.
These recipe examples were tested on October 21, 2025.
Setup#
Import Libraries#
The following external libraries need to be installed into your enviornment to run the code examples in this tutorial:
We import the libraries used in this tutorial below:
import matplotlib.pyplot as plt
import requests
from pprint import pprint
from IPython.display import SVG
from time import sleep
import pandas as pd
from dotenv import load_dotenv
import os
Import API Key#
An API key is required to access the CAS Common Chemistry API. You can sign up for one at the Request API Access for CAS Common Chemistry page.
We keep our API key in a .env file and use the dotenv library to access it. If you would like to use this method, create a .env file and add the following line to it:
CAS_API_KEY=PUT_YOUR_API_KEY_HERE
load_dotenv()
try:
HEADERS = {"X-API-KEY": os.environ["CAS_API_KEY"]}
except KeyError:
print("API key not found. Please set 'CAS_API_KEY' in your .env file.")
1. Common Chemistry Record Detail Retrieval#
A CAS Registry Number (CAS RN) is a unique identifier for a specific substance. Information about substances in CAS Common Chemistry can be retrieved using the /detail endpoint and a CAS RN.
BASE_URL = "https://commonchemistry.cas.org/api/"
endpoint = "detail"
params = {
"cas_rn": "10094-36-7" # Ethyl cyclohexanepropionate
}
response = requests.get(f"{BASE_URL}{endpoint}", params=params, headers=HEADERS)
# Status code 200 indicates success
response.status_code
200
data = response.json()
# Print first level of the response data
pprint(data, depth=1)
{'canonicalSmile': 'O=C(OCC)CCC1CCCCC1',
'experimentalProperties': [...],
'hasMolfile': True,
'images': [...],
'inchi': 'InChI=1S/C11H20O2/c1-2-13-11(12)9-8-10-6-4-3-5-7-10/h10H,2-9H2,1H3',
'inchiKey': 'InChIKey=NRVPMFHPHGBQLP-UHFFFAOYSA-N',
'molecularFormula': 'C<sub>11</sub>H<sub>20</sub>O<sub>2</sub>',
'molecularMass': '184.28',
'name': 'Ethyl cyclohexanepropionate',
'propertyCitations': [...],
'replacedRns': [],
'rn': '10094-36-7',
'smile': 'C(CC(OCC)=O)C1CCCCC1',
'synonyms': [...],
'uri': 'substance/pt/10094367'}
Accessing Specific Data#
We can index into the data dictionary to access data from the response.
# Get Experimental Properties
data["experimentalProperties"][0]
{'name': 'Boiling Point',
'property': '105-113 °C @ Press: 17 Torr',
'sourceNumber': 1}
# Get Boiling Point property
data["experimentalProperties"][0]["property"]
'105-113 °C @ Press: 17 Torr'
# Get InChIKey
data["inchiKey"]
'InChIKey=NRVPMFHPHGBQLP-UHFFFAOYSA-N'
# Get Canonical SMILES
data["canonicalSmile"]
'O=C(OCC)CCC1CCCCC1'
Display the Molecule Drawing#
The images section of the data contains data formatted as Scalable Vector Graphics (SVG). SVG is an XML based vector format for defining two-dimensional graphics and has been an open standard since 1999. Using the SVG data provided by the API, we display the molecule drawing using the SVG() method from IPython.display.
svg_string = data["images"][0]
# Display the molecule
display(SVG(svg_string))
2. Common Chemistry API Record Detail Retrieval in a Loop#
To obtain the data for multiple records, we request data for each CAS RN in a loop and save it to a list.
casrns = ["10094-36-7", "10031-92-2", "10199-61-8", "10036-21-2", "1019020-13-3"]
casrn_data_list = []
for casrn in casrns:
params = {
"cas_rn": casrn
}
try:
response = requests.get(f"{BASE_URL}{endpoint}", params=params, headers=HEADERS)
# Add a delay between API calls
sleep(1)
# Raise an error for bad responses
response.raise_for_status()
casrn_data_list.append(response.json())
except requests.exceptions.RequestException as e:
print(f"An error occurred: {e}")
# View the first item's summary
pprint(casrn_data_list[0], depth=1)
{'canonicalSmile': 'O=C(OCC)CCC1CCCCC1',
'experimentalProperties': [...],
'hasMolfile': True,
'images': [...],
'inchi': 'InChI=1S/C11H20O2/c1-2-13-11(12)9-8-10-6-4-3-5-7-10/h10H,2-9H2,1H3',
'inchiKey': 'InChIKey=NRVPMFHPHGBQLP-UHFFFAOYSA-N',
'molecularFormula': 'C<sub>11</sub>H<sub>20</sub>O<sub>2</sub>',
'molecularMass': '184.28',
'name': 'Ethyl cyclohexanepropionate',
'propertyCitations': [...],
'replacedRns': [],
'rn': '10094-36-7',
'smile': 'C(CC(OCC)=O)C1CCCCC1',
'synonyms': [...],
'uri': 'substance/pt/10094367'}
Select Some Specific Data#
# Get Canonical SMILES
cansmiles = [item["canonicalSmile"] for item in casrn_data_list]
cansmiles
['O=C(OCC)CCC1CCCCC1',
'O=C(C#CCCCCCC)OCC',
'O=C(OCC)CN1N=CC=C1',
'O=C(OCC)C1=CC=CC(=C1)CCC(=O)OCC',
'N=C(OCC)C1=CCCCC1']
# Get Synonyms
synonyms_list = []
for casrn_item in casrn_data_list:
# Since the synonyms are a list, we need to extend the list
# Instead of appending to avoid nested lists
synonyms_list.extend(casrn_item["synonyms"])
synonyms_list
['Cyclohexanepropanoic acid, ethyl ester',
'Cyclohexanepropionic acid, ethyl ester',
'Ethyl cyclohexanepropionate',
'Ethyl cyclohexylpropanoate',
'Ethyl 3-cyclohexylpropionate',
'Ethyl 3-cyclohexylpropanoate',
'3-Cyclohexylpropionic acid ethyl ester',
'NSC 71463',
'Ethyl 3-cyclohexanepropionate',
'2-Nonynoic acid, ethyl ester',
'Ethyl 2-nonynoate',
'NSC 190985',
'1<em>H</em>-Pyrazole-1-acetic acid, ethyl ester',
'Pyrazole-1-acetic acid, ethyl ester',
'Ethyl 1<em>H</em>-pyrazole-1-acetate',
'Ethyl 1-pyrazoleacetate',
'Ethyl 2-(1<em>H</em>-pyrazol-1-yl)acetate',
'Benzenepropanoic acid, 3-(ethoxycarbonyl)-, ethyl ester',
'Hydrocinnamic acid, <em>m</em>-carboxy-, diethyl ester',
'Ethyl 3-(ethoxycarbonyl)benzenepropanoate',
'1-Cyclohexene-1-carboximidic acid, ethyl ester',
'Ethyl 1-cyclohexene-1-carboximidate']
Display Molecule Drawings#
# Get SVG image text
svg_strings = []
for casrn_item in casrn_data_list:
svg_strings.append(casrn_item["images"][0])
# Display the molecules
for svg_string in svg_strings:
display(SVG(svg_string))
Create Pandas DataFrame#
df = pd.json_normalize(casrn_data_list)
# Select specific columns to display
df_subset = df[["uri", "rn", "name", "inchiKey", "canonicalSmile", "molecularMass"]]
df
| uri | rn | name | images | inchi | inchiKey | smile | canonicalSmile | molecularFormula | molecularMass | experimentalProperties | propertyCitations | synonyms | replacedRns | hasMolfile | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | substance/pt/10094367 | 10094-36-7 | Ethyl cyclohexanepropionate | [<svg width="215" viewBox="0 0 215 101" style=... | InChI=1S/C11H20O2/c1-2-13-11(12)9-8-10-6-4-3-5... | InChIKey=NRVPMFHPHGBQLP-UHFFFAOYSA-N | C(CC(OCC)=O)C1CCCCC1 | O=C(OCC)CCC1CCCCC1 | C<sub>11</sub>H<sub>20</sub>O<sub>2</sub> | 184.28 | [{'name': 'Boiling Point', 'property': '105-11... | [{'docUri': 'document/pt/document/22252593', '... | [Cyclohexanepropanoic acid, ethyl ester, Cyclo... | [] | True |
| 1 | substance/pt/10031922 | 10031-92-2 | Ethyl 2-nonynoate | [<svg width="292" viewBox="0 0 292 86" style="... | InChI=1S/C11H18O2/c1-3-5-6-7-8-9-10-11(12)13-4... | InChIKey=BFZNMUGAZYAMTG-UHFFFAOYSA-N | C(C#CCCCCCC)(OCC)=O | O=C(C#CCCCCCC)OCC | C<sub>11</sub>H<sub>18</sub>O<sub>2</sub> | 182.26 | [] | [] | [2-Nonynoic acid, ethyl ester, Ethyl 2-nonynoa... | [] | True |
| 2 | substance/pt/10199618 | 10199-61-8 | Ethyl 1<em>H</em>-pyrazole-1-acetate | [<svg width="184" viewBox="0 0 184 62" style="... | InChI=1S/C7H10N2O2/c1-2-11-7(10)6-9-5-3-4-8-9/... | InChIKey=SEHJVNBWAGPXSM-UHFFFAOYSA-N | C(C(OCC)=O)N1C=CC=N1 | O=C(OCC)CN1N=CC=C1 | C<sub>7</sub>H<sub>10</sub>N<sub>2</sub>O<sub>... | 154.17 | [{'name': 'Boiling Point', 'property': '130 °C... | [{'docUri': 'document/pt/document/17362219', '... | [1<em>H</em>-Pyrazole-1-acetic acid, ethyl est... | [] | True |
| 3 | substance/pt/10036212 | 10036-21-2 | Ethyl 3-(ethoxycarbonyl)benzenepropanoate | [<svg width="318" viewBox="0 0 318 101" style=... | InChI=1S/C14H18O4/c1-3-17-13(15)9-8-11-6-5-7-1... | InChIKey=HDGSRVUQMHJTDH-UHFFFAOYSA-N | C(OCC)(=O)C1=CC(CCC(OCC)=O)=CC=C1 | O=C(OCC)C1=CC=CC(=C1)CCC(=O)OCC | C<sub>14</sub>H<sub>18</sub>O<sub>4</sub> | 250.29 | [] | [] | [Benzenepropanoic acid, 3-(ethoxycarbonyl)-, e... | [] | True |
| 4 | substance/pt/1019020133 | 1019020-13-3 | Ethyl 1-cyclohexene-1-carboximidate | [<svg width="163" viewBox="0 0 163 101" style=... | InChI=1S/C9H15NO/c1-2-11-9(10)8-6-4-3-5-7-8/h6... | InChIKey=OYGZRJWLVOOMBM-UHFFFAOYSA-N | C(OCC)(=N)C=1CCCCC1 | N=C(OCC)C1=CCCCC1 | C<sub>9</sub>H<sub>15</sub>NO | 153.22 | [] | [] | [1-Cyclohexene-1-carboximidic acid, ethyl este... | [] | True |
3. Common Chemistry Search#
In addition to the /detail API, the CAS Common Chemistry API has a /search method that allows searching by CAS RN, SMILES, InChI/InChIKey, and name.
endpoint = "search"
params = {
# InChIKey for Quinine
"q": "InChIKey=LOUPRKONTZGTKE-WZBLMQSHSA-N",
}
try:
response = requests.get(f"{BASE_URL}{endpoint}", params=params, headers=HEADERS)
# Raise an error for bad responses
response.raise_for_status()
data = response.json()
pprint(data, depth=3)
except requests.exceptions.RequestException as e:
print(f"An error occurred: {e}")
data = None
{'count': 1,
'results': [{'images': [...], 'name': 'Quinine', 'rn': '130-95-0'}]}
Note that with the CAS Common Chemistry Search API, only the image data, name, and CAS RN is returned. In order to retrieve the full record, we can combine our search with the related detail API:
# Extract CAS RN
quinine_rn = data["results"][0]["rn"]
quinine_rn
'130-95-0'
# Get detailed record for quinine
endpoint = "detail"
params = {
# Quinine CAS RN
"cas_rn": quinine_rn
}
response = requests.get(f"{BASE_URL}{endpoint}", params=params, headers=HEADERS)
data = response.json()
pprint(data, depth=1)
{'canonicalSmile': 'OC(C=1C=CN=C2C=CC(OC)=CC21)C3N4CCC(C3)C(C=C)C4',
'experimentalProperties': [...],
'hasMolfile': True,
'images': [...],
'inchi': 'InChI=1S/C20H24N2O2/c1-3-13-12-22-9-7-14(13)10-19(22)20(23)16-6-8-21-18-5-4-15(24-2)11-17(16)18/h3-6,8,11,13-14,19-20,23H,1,7,9-10,12H2,2H3/t13-,14-,19-,20+/m0/s1',
'inchiKey': 'InChIKey=LOUPRKONTZGTKE-WZBLMQSHSA-N',
'molecularFormula': 'C<sub>20</sub>H<sub>24</sub>N<sub>2</sub>O<sub>2</sub>',
'molecularMass': '324.42',
'name': 'Quinine',
'propertyCitations': [...],
'replacedRns': [...],
'rn': '130-95-0',
'smile': '[C@@H](O)(C=1C2=C(C=CC(OC)=C2)N=CC1)[C@]3([N@@]4C[C@H](C=C)[C@](C3)(CC4)[H])[H]',
'synonyms': [...],
'uri': 'substance/pt/130950'}
Handle Multiple Results#
# Set up search query parameters
endpoint = "search"
params = {
# SMILES for butadiene
"q": "C=CC=C"
}
# Request data from CAS Common Chemistry Search API
response = requests.get(f"{BASE_URL}{endpoint}", params=params, headers=HEADERS)
# Status code 200 indicates success
response.status_code
200
data = response.json()
# Get results count
data["count"]
7
# Extract out CAS RNs
smi_casrn_list = []
for item in data["results"]:
smi_casrn_list.append(item["rn"])
smi_casrn_list
['106-99-0',
'16422-75-6',
'26952-74-9',
'29406-96-0',
'29989-19-3',
'31567-90-5',
'9003-17-2']
# Now use the detail API to retrieve the full records
endpoint = "detail"
smi_detail_data = []
for casrn in smi_casrn_list:
params = {
"cas_rn": casrn
}
try:
response = requests.get(f'{BASE_URL}{endpoint}', params=params, headers=HEADERS)
# Add a delay between API calls
sleep(1)
# Raise an error for bad responses
response.raise_for_status()
smi_detail_data.append(response.json())
except requests.exceptions.RequestException as e:
print(f"An error occurred: {e}")
# Get names from the detail records
names = [item["name"] for item in smi_detail_data]
names
['1,3-Butadiene',
'Butadiene trimer',
'Butadiene dimer',
'1,3-Butadiene, homopolymer, isotactic',
'1,3-Butadiene-<em>1</em>,<em>1</em>,<em>2</em>,<em>3</em>,<em>4</em>,<em>4</em>-<em>d</em><sub>6</sub>, homopolymer',
'Syndiotactic polybutadiene',
'Polybutadiene']
Handle Multiple Page Results#
The CAS Common Chemistry API returns 50 results per page, and only the first page is returned by default. If the search returns more than 50 results, the offset option can be added to page through and obtain all results:
# Set up search query parameters
endpoint = "search"
params = {
"q": "selen*"
}
# Get results count for CAS Common Chemistry Search
response = requests.get(f"{BASE_URL}{endpoint}", params=params, headers=HEADERS)
# Status code 200 indicates success
response.status_code
200
data = response.json()
num_results = data["count"]
num_results
194
# Request data and save to a list in a loop for each page
search_data_list = []
# Split the search into pages of 50 results each
for page_idx in range(int(num_results/50 + 1)):
params = {
"q": "selen*",
"offset": page_idx * 50
}
try:
response = requests.get(BASE_URL + endpoint, params=params, headers=HEADERS)
# Add a delay between API calls
sleep(1)
# Raise an error for bad responses
response.raise_for_status()
search_data_list.extend(response.json()['results'])
except requests.exceptions.RequestException as e:
print(f"An error occurred: {e}")
len(search_data_list)
194
# We can index and extract out the first casrn like this
search_data_list[0]["rn"]
'10025-68-0'
# Extract out all CAS RNs from the list of lists
casrn_list = [item["rn"] for item in search_data_list]
# Show first 10
casrn_list[:10]
['10025-68-0',
'10026-03-6',
'10026-23-0',
'10101-96-9',
'10102-18-8',
'10102-23-5',
'10112-94-4',
'10161-84-9',
'10214-40-1',
'10236-58-5']
# Now we can loop through each casrn and use the detail API to obtain the entire record
# This will take about 5 minutes
endpoint = "detail"
detail_data_list = []
for casrn in casrn_list:
params = {
"cas_rn": casrn
}
try:
response = requests.get(f'{BASE_URL}{endpoint}', params=params, headers=HEADERS)
# Add a delay between API calls
sleep(1)
# Raise an error for bad responses
response.raise_for_status()
detail_data_list.append(response.json())
except requests.exceptions.RequestException as e:
print(f"An error occurred: {e}")
# Extract out some data such as molecularMass
mms = [item["molecularMass"] for item in detail_data_list]
# Print number of molecular masses retrieved
len(mms)
194
# View first 20
# Note that several do not have molecularMass values and have an empty string in the record
mms[:20]
['228.82',
'220.77',
'187.91',
'187.67',
'174.95',
'371.11',
'167.96',
'300.24',
'192.52',
'168.05',
'410.78',
'207.17',
'586.23',
'431.38',
'167.20',
'241.11',
'192.07',
'368.25',
'265.00',
'']
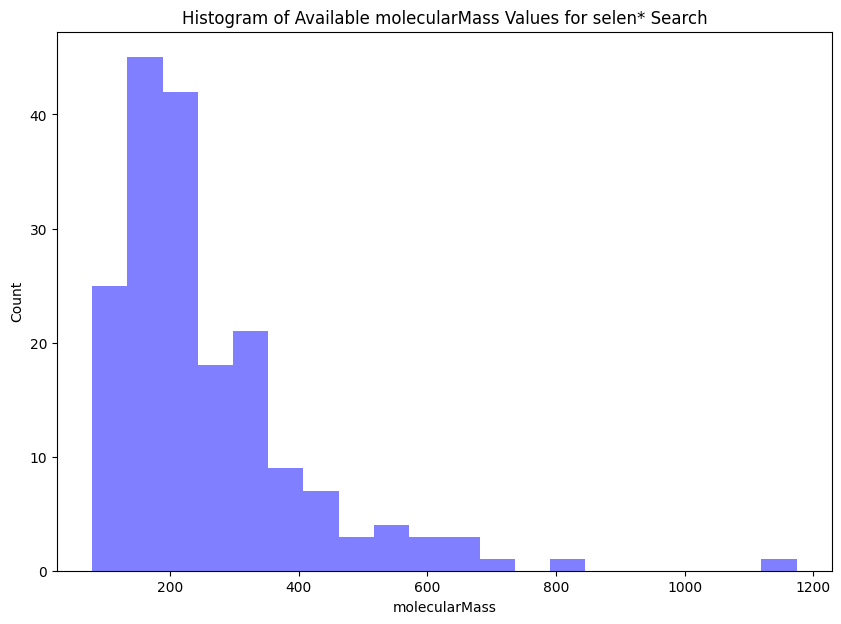
# Finally, we can even quickly create a simple visualization from the
# Extracted molecularMass values (from the selen* search)
# Remove empty strings
mms_values = list(filter(None, mms))
# Convert to floats
mms_values_float = [float(mms_value) for mms_value in mms_values]
# Plot data
plt.figure(figsize=(10,7))
plt.hist(mms_values_float, histtype='bar',bins = 20, facecolor="blue", alpha=0.5)
plt.title("Histogram of Available molecularMass Values for selen* Search")
plt.xlabel("molecularMass")
plt.ylabel("Count")
plt.show()