PubChem API in Python#
By Avery Fernandez
PubChem provides programmatic access to chemical data and bioactivity information from the National Center for Biotechnology Information (NCBI), enabling efficient retrieval and analysis of chemical structures, identifiers, properties, and associated biological activities.
Please see the following resources for more information on API usage:
Documentation
Terms
Data Reuse
NOTE: The PubChem limits requests to a maximum of 5 requests per second.
These recipe examples were tested on April 9, 2025.
Attribution: This tutorial was adapted from supporting information in:
Scalfani, V. F.; Ralph, S. C. Alshaikh, A. A.; Bara, J. E. Programmatic Compilation of Chemical Data and Literature From PubChem Using Matlab. Chemical Engineering Education, 2020, 54, 230. https://doi.org/10.18260/2-1-370.660-115508 and vfscalfani/MATLAB-cheminformatics
Setup#
Import Libraries#
The following external libraries need to be installed into your environment to run the code examples in this tutorial:
We import the libraries used in this tutorial below:
import requests
from pprint import pprint
from time import sleep
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
1. PubChem Similarity#
Get Compound Image#
We can search for a compound and display an image, for example: 1-Butyl-3-methyl-imidazolium; CID = 2734162
# Request PNG from PubChem and save file
base_url = "https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/"
compoundID = "2734162"
try:
response = requests.get(base_url + "cid/" + compoundID + "/PNG")
response.raise_for_status()
img = response.content
with open(f"{compoundID}.png", "wb") as out:
out.write(img)
except requests.exceptions.RequestException as e:
print(f"Error fetching PNG for CID {compoundID}: {e}")
# Display compound PNG with Matplotlib
img = mpimg.imread(f"{compoundID}.png")
plt.imshow(img)
plt.show()
Retrieve InChI and SMILES#
properties = ["IsomericSMILES", "InChI"]
try:
response = requests.get(
base_url + "cid/" + compoundID + f"/property/{','.join(properties)}/JSON"
)
response.raise_for_status()
data = response.json()
except requests.exceptions.RequestException as e:
print(f"Error fetching properties for CID {compoundID}: {e}")
data = None
if data:
pprint(data)
{'PropertyTable': {'Properties': [{'CID': 2734162,
'InChI': 'InChI=1S/C8H15N2/c1-3-4-5-10-7-6-9(2)8-10/h6-8H,3-5H2,1-2H3/q+1',
'IsomericSMILES': 'CCCCN1C=C[N+](=C1)C'}]}}
# Extract InChI
if data:
print("Extracted InChI:")
print(data["PropertyTable"]["Properties"][0]["InChI"])
Extracted InChI:
InChI=1S/C8H15N2/c1-3-4-5-10-7-6-9(2)8-10/h6-8H,3-5H2,1-2H3/q+1
# Extract Isomeric SMILES
if data:
print("Extracted Isomeric SMILES:")
print(data["PropertyTable"]["Properties"][0]["IsomericSMILES"])
Extracted Isomeric SMILES:
CCCCN1C=C[N+](=C1)C
Perform a Similarity Search#
We will use the PubChem API to perform a Fingerprint Tanimoto Similarity Search (SS).
(2D Tanimoto threshold 95% to 1-Butyl-3-methyl-imidazolium; CID = 2734162)
try:
response = requests.get(
base_url + "fastsimilarity_2d/cid/" + compoundID + "/cids/JSON?Threshold=95"
)
response.raise_for_status()
data = response.json()
except requests.exceptions.RequestException as e:
print(f"Error fetching similar compounds for CID {compoundID}: {e}")
data = None
if data:
id_list = data["IdentifierList"]["CID"]
else:
id_list = []
In the above request value, you can adjust to the desired Tanimoto threshold (i.e., 97, 90, etc.)
len(id_list)
293
# display first 25
id_list[0:25]
[61347,
529334,
2734161,
118785,
12971008,
2734162,
11171745,
11424151,
11448496,
304622,
2734236,
11160028,
20148470,
11245926,
87560886,
87754289,
2734168,
5245884,
53384410,
87942618,
4183883,
10313448,
10537570,
11788435,
15557008]
Retrieve Identifier and Property Data#
Get the following data for the retrieved CIDs (idList): InChI, Isomeric SMILES, MW, Heavy Atom Count, Rotable Bond Count, and Charge
compoundDictionary = []
properties = ["InChI", "IsomericSMILES", "MolecularWeight",
"HeavyAtomCount", "RotatableBondCount", "Charge"]
for cid in id_list:
try:
response = requests.get(
base_url + "cid/" + str(cid) + f"/property/{','.join(properties)}/JSON"
)
sleep(.25)
response.raise_for_status()
data = response.json()
compoundDictionary.append(data["PropertyTable"]["Properties"][0])
except requests.exceptions.RequestException as e:
print(f"Error fetching properties for CID {cid}: {e}")
len(compoundDictionary)
293
pprint(compoundDictionary[0:3])
[{'CID': 61347,
'Charge': 0,
'HeavyAtomCount': 9,
'InChI': 'InChI=1S/C7H12N2/c1-2-3-5-9-6-4-8-7-9/h4,6-7H,2-3,5H2,1H3',
'IsomericSMILES': 'CCCCN1C=CN=C1',
'MolecularWeight': '124.18',
'RotatableBondCount': 3},
{'CID': 529334,
'Charge': 0,
'HeavyAtomCount': 10,
'InChI': 'InChI=1S/C8H14N2/c1-2-3-4-6-10-7-5-9-8-10/h5,7-8H,2-4,6H2,1H3',
'IsomericSMILES': 'CCCCCN1C=CN=C1',
'MolecularWeight': '138.21',
'RotatableBondCount': 4},
{'CID': 2734161,
'Charge': 0,
'HeavyAtomCount': 11,
'InChI': 'InChI=1S/C8H15N2.ClH/c1-3-4-5-10-7-6-9(2)8-10;/h6-8H,3-5H2,1-2H3;1H/q+1;/p-1',
'IsomericSMILES': 'CCCCN1C=C[N+](=C1)C.[Cl-]',
'MolecularWeight': '174.67',
'RotatableBondCount': 3}]
Data Table#
We can display the dictionary as a data table, but we will only do this for the first 25:
# numbers in print statement indicate amount of space used
print ("{:<10} {:<8} {:<16} {:<25} {:<40} {:<18} {:<4} ".format("CID", "Charge",
"HeavyAtomCount", "InChI", "IsomericSMILES", "MolecularWeight", "RotatableBondCount"))
for compound in compoundDictionary[0:25]:
cid = compound["CID"]
charge = compound["Charge"]
heavyAtom = compound["HeavyAtomCount"]
# only display first 30 characters of InChI
inchi = compound["InChI"][0:20] + "..."
isomeric = compound["IsomericSMILES"]
molecular = compound["MolecularWeight"]
rotatable = compound["RotatableBondCount"]
print ("{:<10} {:<8} {:<16} {:<25} {:<40} {:<18} {:<4} ".format(cid, charge, heavyAtom,
inchi, isomeric, molecular, rotatable))
CID Charge HeavyAtomCount InChI IsomericSMILES MolecularWeight RotatableBondCount
61347 0 9 InChI=1S/C7H12N2/c1-... CCCCN1C=CN=C1 124.18 3
529334 0 10 InChI=1S/C8H14N2/c1-... CCCCCN1C=CN=C1 138.21 4
2734161 0 11 InChI=1S/C8H15N2.ClH... CCCCN1C=C[N+](=C1)C.[Cl-] 174.67 3
118785 0 8 InChI=1S/C6H10N2/c1-... CCCN1C=CN=C1 110.16 2
12971008 0 10 InChI=1S/C7H13N2.HI/... CCCN1C=C[N+](=C1)C.[I-] 252.10 2
2734162 1 10 InChI=1S/C8H15N2/c1-... CCCCN1C=C[N+](=C1)C 139.22 3
11171745 0 15 InChI=1S/C8H15N2.C2N... CCCCN1C=C[N+](=C1)C.C(=[N-])=NC#N 205.26 3
11424151 0 13 InChI=1S/C8H15N2.CHN... CCCCN1C=C[N+](=C1)C.C(#N)[S-] 197.30 3
11448496 0 11 InChI=1S/C8H15N2.HI/... CCCCN1C=C[N+](=C1)C.[I-] 266.12 3
304622 0 10 InChI=1S/C8H14N2/c1-... CCCCN1C=CN=C1C 138.21 3
2734236 0 11 InChI=1S/C8H15N2.BrH... CCCCN1C=C[N+](=C1)C.[Br-] 219.12 3
11160028 0 10 InChI=1S/C7H13N2.BrH... CCCN1C=C[N+](=C1)C.[Br-] 205.10 2
20148470 0 10 InChI=1S/C8H14N2/c1-... CC(C)CCN1C=CN=C1 138.21 3
11245926 0 13 InChI=1S/C8H15N2.Br2... CCCCN1C=C[N+](=C1)C.[Br-].BrBr 378.93 3
87560886 0 12 InChI=1S/C9H15N2.BrH... CCCC[N+]1=CN(C=C1)C=C.[Br-] 231.13 4
87754289 0 12 InChI=1S/C9H17N2.BrH... CCCC[N+]1=CN(C=C1)CC.[Br-] 233.15 4
2734168 1 11 InChI=1S/C9H17N2/c1-... CCCCN1C=C[N+](=C1C)C 153.24 3
5245884 1 9 InChI=1S/C7H13N2/c1-... CCCN1C=C[N+](=C1)C 125.19 2
53384410 0 13 InChI=1S/C8H15N2.Br3... CCCCN1C=C[N+](=C1)C.Br[Br-]Br 378.93 3
87942618 0 11 InChI=1S/C8H15N2.BrH... CCC[N+]1=CN(C=C1)CC.[Br-] 219.12 3
4183883 1 10 InChI=1S/C8H15N2/c1-... CCCN1C=C[N+](=C1C)C 139.22 2
10313448 1 11 InChI=1S/C9H17N2/c1-... CCCC[N+]1=CN(C=C1)CC 153.24 4
10537570 1 11 InChI=1S/C9H17N2/c1-... CCCCCN1C=C[N+](=C1)C 153.24 4
11788435 0 11 InChI=1S/C8H15N2.H2O... CCCCN1C=C[N+](=C1)C.[OH-] 156.23 3
15557008 0 10 InChI=1S/C8H14N2/c1-... CCCC1=NC=CN1CC 138.21 3
Retrieve Images of Compounds from Similarity Search#
# we will only do this for the first five:
for cid in id_list[0:5]:
try:
response = requests.get(base_url + "cid/" + str(cid) + "/PNG")
sleep(.25)
response.raise_for_status()
img = response.content
with open(f"{cid}.png", "wb") as out:
out.write(img)
except requests.exceptions.RequestException as e:
print(f"Error fetching PNG for CID {cid}: {e}")
continue
print(cid)
img = mpimg.imread(str(cid) + ".png")
plt.imshow(img)
plt.show()
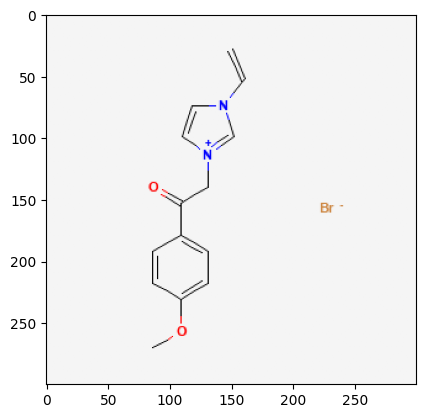
61347
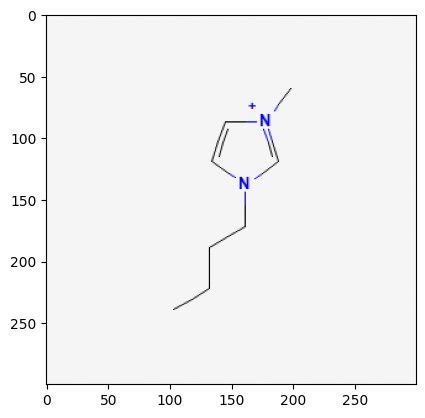
529334
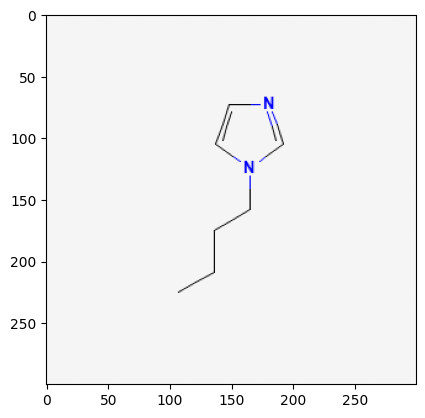
2734161
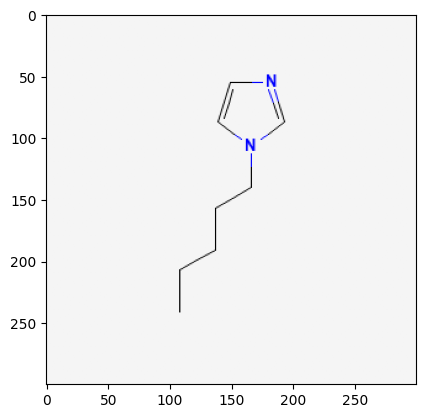
118785
12971008
2. PubChem SMARTS Search#
Search for chemical structures from a SMARTS substructure query.
Define SMARTS Queries#
View pattern syntax at SMARTSPlus.
Note: These are vinyl imidazolium substructure searches
smartsQ = [
"[CR0H2][n+]1[cH1][cH1]n([CR0H1]=[CR0H2])[cH1]1",
"[CR0H2][n+]1[cH1][cH1]n([CR0H2][CR0H1]=[CR0H2])[cH1]1",
"[CR0H2][n+]1[cH1][cH1]n([CR0H2][CR0H2][CR0H1]=[CR0H2])[cH1]1"
]
Add your own SMARTS queries to customize. You can add as many as desired within a list
Perform a SMARTS query search#
combinedList = []
for smarts in smartsQ:
try:
response = requests.get(base_url + "fastsubstructure/smarts/" + smarts + "/cids/JSON")
sleep(.25)
response.raise_for_status()
data = response.json()
combinedList += data["IdentifierList"]["CID"]
except requests.exceptions.RequestException as e:
print(f"Error fetching substructure for SMARTS {smarts}: {e}")
request = None
len(combinedList)
945
pprint(combinedList[0:25]) # display 25
[2881855,
23724184,
2881236,
2881558,
2881232,
2881324,
2881449,
2881640,
24766550,
87327009,
87560886,
87575063,
121235111,
2881597,
2881807,
23724198,
132274871,
1552916,
1912201,
2264809,
46178576,
86657882,
129850195,
129852976,
129853221]
Retrieve Identifier and Property Data#
smartsList = []
properties = ["InChI", "IsomericSMILES", "MolecularWeight",
"IUPACName", "HeavyAtomCount", "CovalentUnitCount", "Charge"]
# demo for first 5 CIDs
for cid in combinedList[0:5]:
try:
response = requests.get(
base_url + "cid/" + str(cid) + f"/property/{','.join(properties)}/JSON"
)
sleep(.25)
response.raise_for_status()
data = response.json()
smartsList.append(data["PropertyTable"]["Properties"][0])
except requests.exceptions.RequestException as e:
print(f"Error fetching properties for CID {cid}: {e}")
pprint(smartsList[0:3])
[{'CID': 2881855,
'Charge': 0,
'CovalentUnitCount': 2,
'HeavyAtomCount': 21,
'IUPACName': '1-(3,4-dimethoxyphenyl)-2-(3-ethenylimidazol-1-ium-1-yl)ethanone;bromide',
'InChI': 'InChI=1S/C15H17N2O3.BrH/c1-4-16-7-8-17(11-16)10-13(18)12-5-6-14(19-2)15(9-12)20-3;/h4-9,11H,1,10H2,2-3H3;1H/q+1;/p-1',
'IsomericSMILES': 'COC1=C(C=C(C=C1)C(=O)C[N+]2=CN(C=C2)C=C)OC.[Br-]',
'MolecularWeight': '353.21'},
{'CID': 23724184,
'Charge': 0,
'CovalentUnitCount': 2,
'HeavyAtomCount': 17,
'IUPACName': '1-(5-bromothiophen-2-yl)-2-(3-ethenylimidazol-1-ium-1-yl)ethanone;bromide',
'InChI': 'InChI=1S/C11H10BrN2OS.BrH/c1-2-13-5-6-14(8-13)7-9(15)10-3-4-11(12)16-10;/h2-6,8H,1,7H2;1H/q+1;/p-1',
'IsomericSMILES': 'C=CN1C=C[N+](=C1)CC(=O)C2=CC=C(S2)Br.[Br-]',
'MolecularWeight': '378.08'},
{'CID': 2881236,
'Charge': 0,
'CovalentUnitCount': 2,
'HeavyAtomCount': 19,
'IUPACName': '1-(3,4-dichlorophenyl)-2-(3-ethenylimidazol-1-ium-1-yl)ethanone;bromide',
'InChI': 'InChI=1S/C13H11Cl2N2O.BrH/c1-2-16-5-6-17(9-16)8-13(18)10-3-4-11(14)12(15)7-10;/h2-7,9H,1,8H2;1H/q+1;/p-1',
'IsomericSMILES': 'C=CN1C=C[N+](=C1)CC(=O)C2=CC(=C(C=C2)Cl)Cl.[Br-]',
'MolecularWeight': '362.0'}]
Retrieve Images of CID Compounds from SMARTS Search#
for cid in combinedList[0:5]:
try:
response = requests.get(base_url + "cid/" + str(cid) + "/PNG")
sleep(.25)
response.raise_for_status()
img = response.content
with open(f"{cid}.png", "wb") as out:
out.write(img)
except requests.exceptions.RequestException as e:
print(f"Error fetching PNG for CID {cid}: {e}")
continue
# display image
pprint(cid)
img = mpimg.imread(str(cid) + ".png")
plt.imshow(img)
plt.show()
2881855
23724184
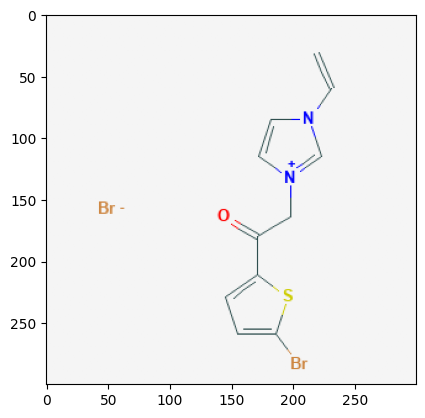
2881236
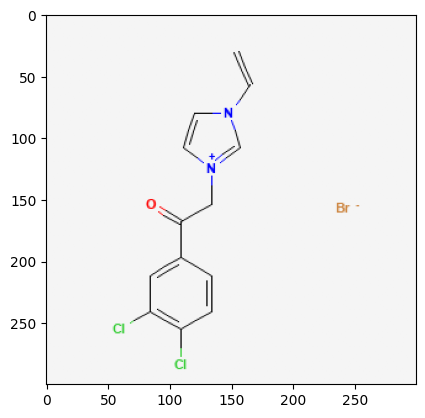
2881558
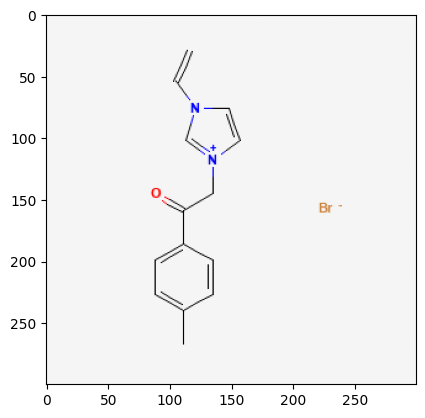
2881232